3X-KBank
Bio BigData Integration Platform
Collect, refine, and integrate a variety of biodata, including genes, drugs, and diseases to present new Insight
We have built a large-scale bio-big data-based in-house platform. By applying AI technology, we can conduct research on the discovery of new drug candidates and various new materials, and have know-how to build and utilize new bio-big data
Introduction
Uncovering obscured relationships within data requires the fusion of diverse databases encompassing genes, transcripts, proteins, and gene expression. This integration offersa panoramic view of intricate biological dynamics. Genes outline blueprints, transcripts illuminate activity, proteins execute functions, and gene expression data unveil regulation nuances. By harmonizing these databases, a multifaceted understanding emerges, illuminating hidden synergies and fueling advances across genetics, molecular biology, and systems analysis.
Challenge
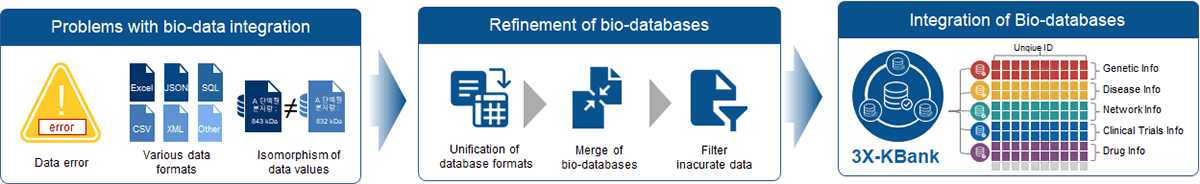
The challenge at hand revolves around the diverse formats and naming conventions within public databases, encompassing Excel, CSV, and Text, along with variations in terminologies. Converting this heterogeneous array of data structures into a cohesive, homogeneous format poses a central challenge. The intricacies of reconciling dissimilar formats, aligning terminologies, and standardizing entries demand meticulous efforts to ensure seamless integration.
Furthermore, the absence of cohesive links compounds the complexity. Establishing connections between disparate datasets, often riddled with distinct terminologies, introduces the additional hurdle of locating and rectifying missing links. This dual challenge requires the formulation of robust methodologies that facilitate the harmonization of diverse data structures, while simultaneously identifying, rectifying, and creating the necessary links to establish a coherent and comprehensive data landscape. Overcoming these challenges is pivotal for harnessing the collective potential of heterogeneous data sources and unlocking valuable insights that transcend the limitations posed by differing formats and missing links.
Solutions
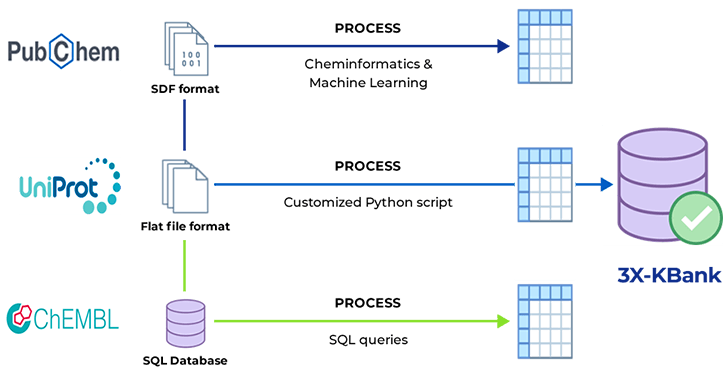
In addressing the intricate challenge of harmonizing diverse biological databases, 3BIGS strategically devised an encompassing solution. Leveraging in-house expertise, they engineered an array of purpose-built scripts, meticulously tailored to parse and standardize data. This multifaceted approach not only transformed varied formats like Excel, CSV, and Text into a cohesive structure but also established intricate links between datasets, transcending disparities in terminology and nomenclature. To streamline data mapping and retrieval, 3BIGS ingeniously renamed data columns with their distinct nomenclature. In pursuit of data integrity, manual curation was employed to rectify missing entries, bolstering the accuracy of the unified dataset. Ultimately, these synergistic efforts coalesced to foster a coherent and comprehensive data landscape, transcending the limitations posed by varied formats and disconnected data sources. The 3BIGS approach exemplifies the fusion of technical innovation and meticulous curation to unlock obscured biological insights and advance the realms of genetics, molecular biology, and systems analysis.
Refinement of Bio-databases
As bio-big data grows, the value of utilization increases when real-world data is refined and quality is selected.
Integration of Bio-databases
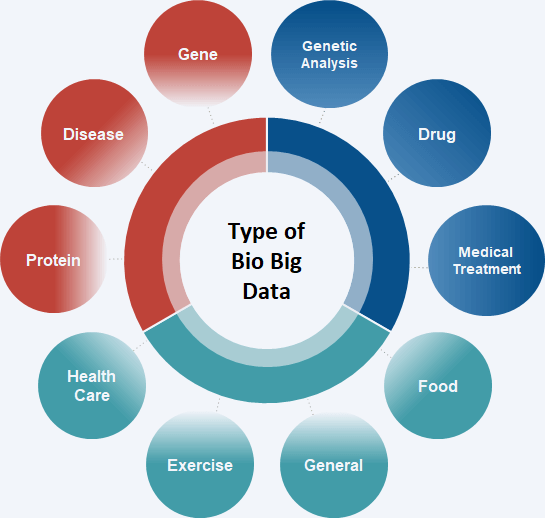
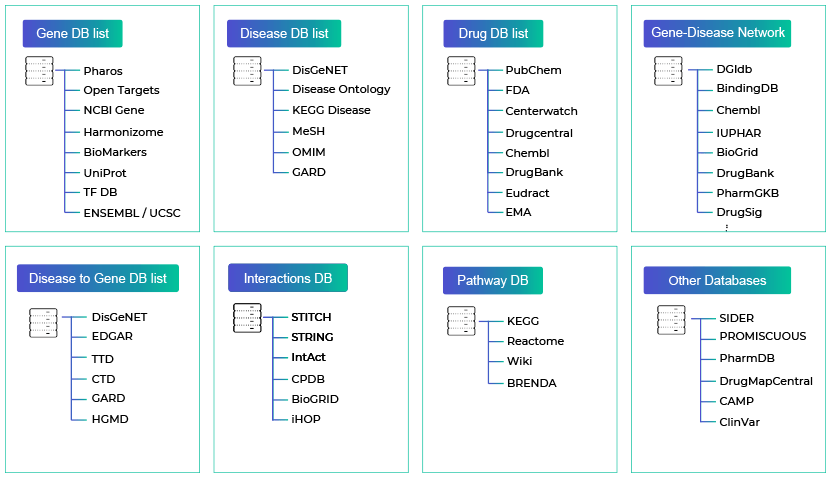
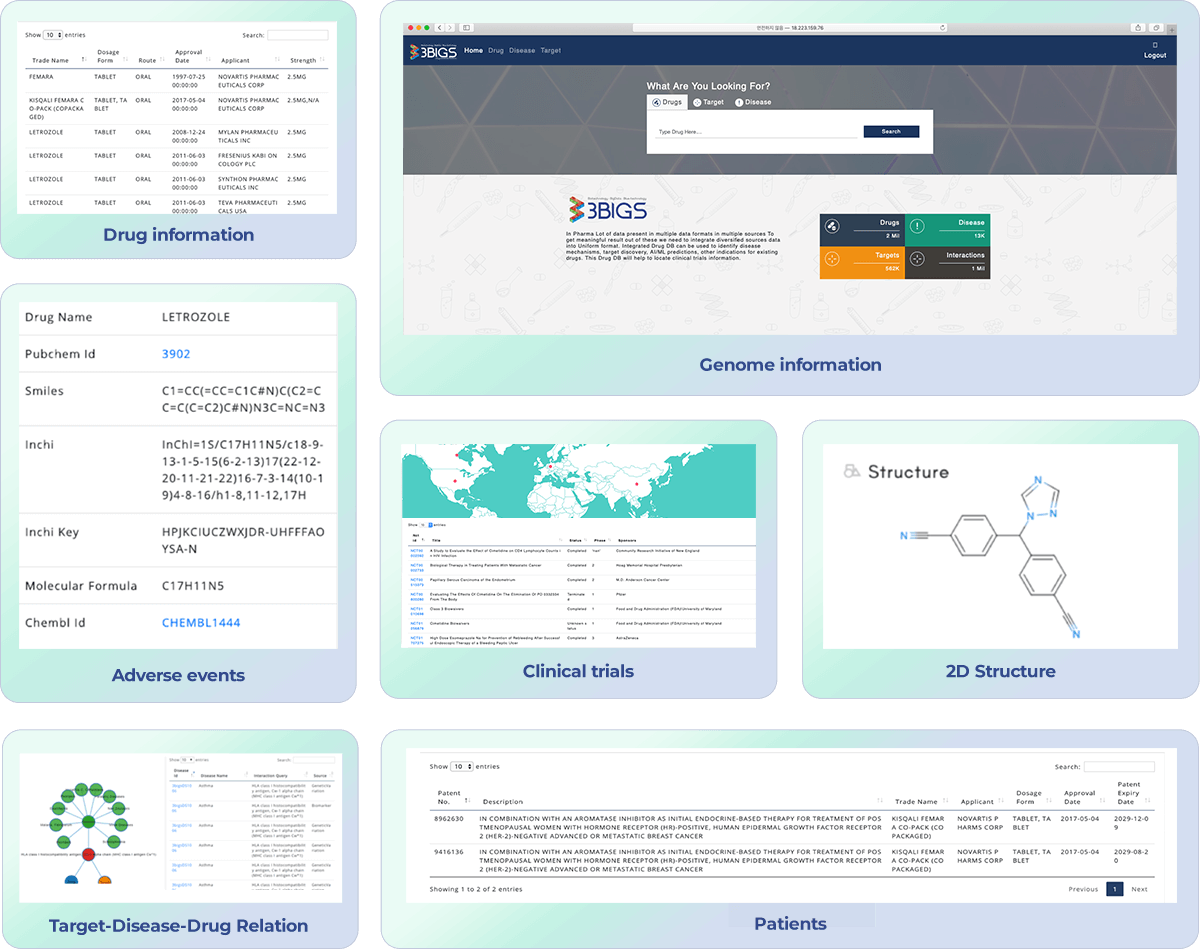
We have collected and refined various data that can be used in bioinformatics, such as more than 70 genes, diseases, drugs, clinical trials, and so on to be used for research purposes.
Utilization of Bio-database
The integrated bio-database can easily collect relevant information such as biomarker discovery, new drug development, gene disease-drug interaction, and supports various analysis results interpretation.
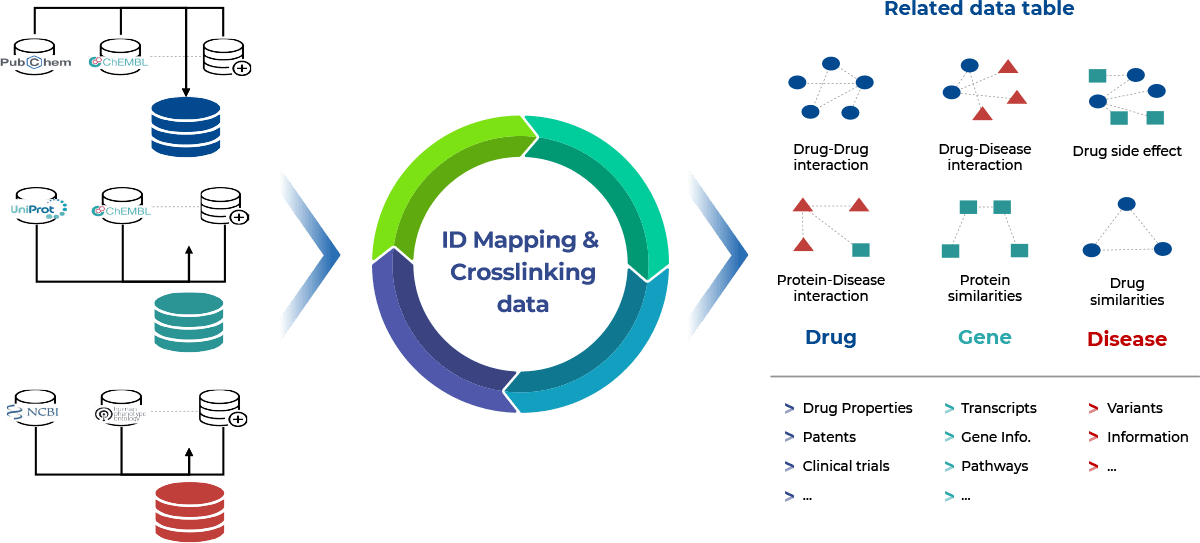
Structure and utilization of scientific data
Based on more than 30 million scientific literature, it structures genes, diseases, drugs, clinical information, and supports various information to identify the flow of research.

 ENGLISH
ENGLISH KOREAN
KOREAN