DNA methylation is analyzed whether a specific gene’s function is expressed by external environmental factors to provide results.
Using the NGS technique, we analyze the methylation of representative DNA during epigenetic regulatory mechanisms.
Analysis Services Features
- Methylation Statistical Analysis
- Visualization of methylation by sample and group
- Group Comparison Analysis
- Analysis report in HTML format
Data Analysis
- MultiQC report
- Alignment Statistics
- Statistical analysis
- Differential Methylation annotation
- Methylation Group analysis
Epigenome Analysis
The epigenome constitutes a vast number of factors that can cause heritable changes through altered gene expression without any changes at the sequence level. Its analysis allows us to decode the intricate relationship we have with our environment. At 3BIGS, our goal is to help you achieve this understanding through leading technologies: CHIP-Seq, ATAC-Seq,
Specialized epigenome analysis we offer:
- Shotgun Metagenomics
- ATAC-seq data analysis
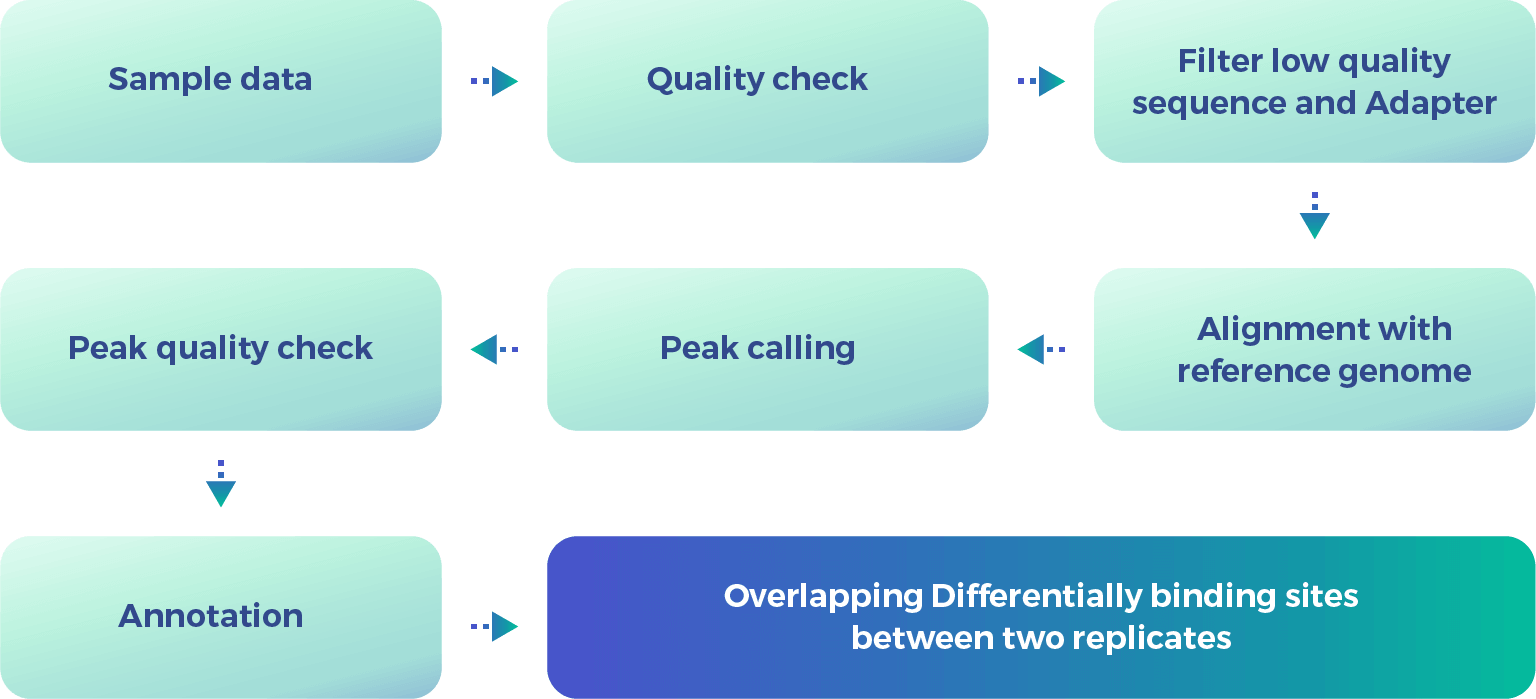
CHIP-Seq Data Analysis
Chromatin Immunoprecipitation Sequencing (CHIP-Seq) is a popular technique utilized for genome-wide characterization of DNA-binding proteins, histone modifications, and nucleosomes. These findings help us discover more about gene regulation and epigenetic mechanisms. 3BIGS has a systematic workflow supported by novel reliable technology designed to cover all your CHIP-Seq data analysis requirements
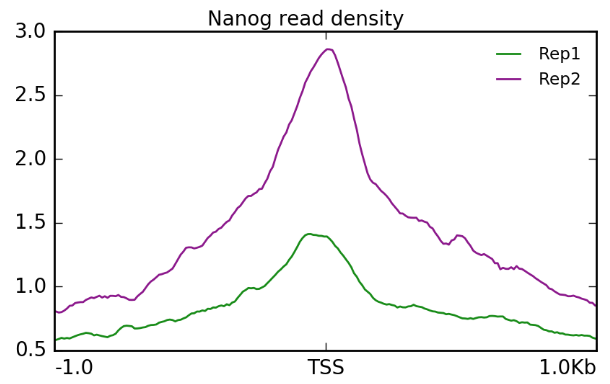
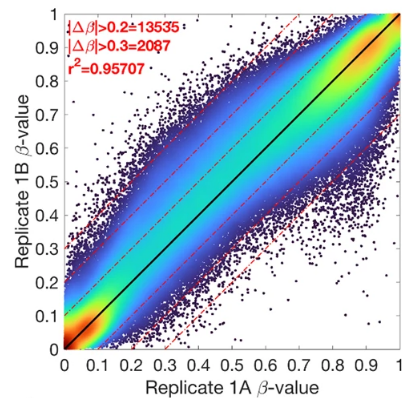
ATAC-Seq Data Analysis
Assay for transposase-accessible chromatin with sequencing (ATAC-Seq) is a widely utilized technique for genome-wide mapping of chromatin accessibility, which can help understand the changes it goes through during abnormal or disease conditions. The 3BIGS workflow effortlessly and reliably gets you one step closer to generating valuable insights regarding the presence of known and unknown regulatory elements crucial for disease management.

 ENGLISH
ENGLISH KOREAN
KOREAN